# A tibble: 3 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.Joining several data frames with dplyr
2025-08-09
Join tables by a shared column
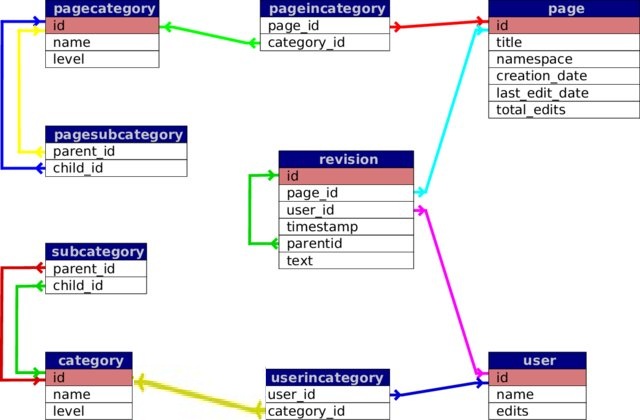
Revision performed by a user
gapminder
geo
geo <- read_csv(glue("https://raw.githubusercontent.com/open-numbers/",
"ddf--gapminder--fasttrack/master/",
"ddf--entities--geo--country.csv"))
geo <- geo %>% select(country, name, main_religion_2008, income_3groups, world_4region)
glimpse(geo)Rows: 273
Columns: 5
$ country <chr> "abkh", "abw", "afg", "ago", "aia", "akr_a_dhe", "a…
$ name <chr> "Abkhazia", "Aruba", "Afghanistan", "Angola", "Angu…
$ main_religion_2008 <chr> NA, "christian", "muslim", "christian", "christian"…
$ income_3groups <chr> NA, "high_income", "low_income", "middle_income", N…
$ world_4region <chr> "europe", "americas", "asia", "africa", "americas",…Now we have a data frame called geo. It lists 273 countries, each only once, and for each country it gives us its dominant religion (snapshot taken in 2008) and to which income group it belongs. The income groups are Gapminder’s own design and obviously refer to the time of creating this dataset. The countries are encoded by their names (column name) and by the abbreviations of these names (column country).
Important
The gapminder data frame does not use abbreviations for country names. But even more importantly, its country column does not correspond to country but to name in the geo data frame. This is something to keep in mind when we try to join these two tables!
Countries in geo vs. gapminder
When we want to join geo and gapminder, we can apparently use geo’s name and gapminder’s country as the primary key - foreign key pair.
The first question you must ask when you want to join two tables by a pair of keys is how much overlap they have at all. geo contains many more countries than gapminder, so much is clear. But it is not given that all of its countries are listed in geo! Therefore we do this quick check.
Note
You could as well call distinct and pull on each, but this base-R way is shorter. It accesses a data frame column as a vector and calls the unique function, which acts on vectors like dplyr::distinct on data frames.
Little overlap in key column values?
[1] "Korea, Dem. Rep." "Korea, Rep." "Swaziland"
[4] "United Kingdom" "United States" "West Bank and Gaza"
[7] "Yemen, Rep." setdiff tells you which elements of the first vectors are not in the second vector. We see that the gapminder data frame is not a full subset of the geo data frame. When we enrich gapminder with the information from geo, some values will be missing. This has no programmatic solution. This is your design decision. Drop these seven countries from gapminder because you don’t get the additional information, or live with NAs in the new columns? Or you even find the geo table more important and only want to enrich with gapminder’s pop, lifeExp, and gdpPercap in a given year? For each of your possible decisions, dplyr comes with a dedicated _join function.
Control key selection
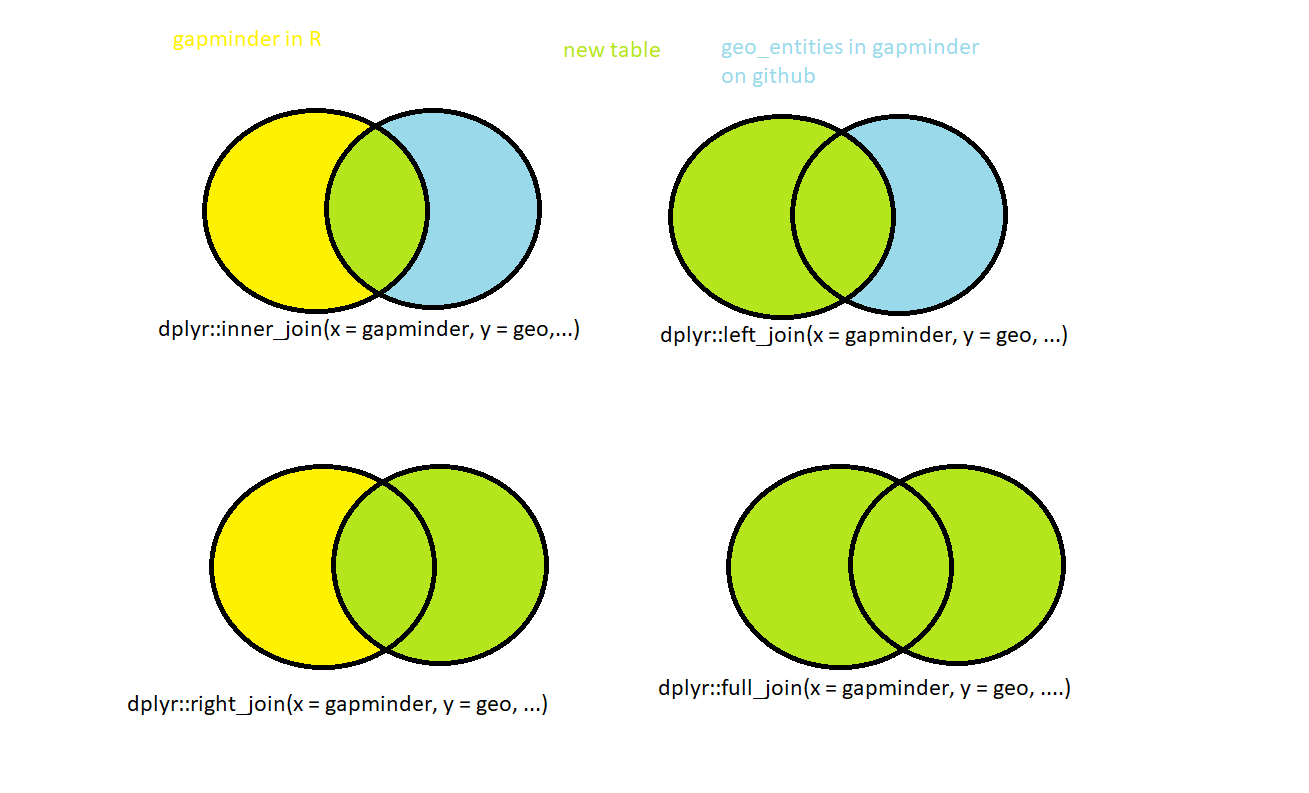
When blue and yellow mix, they give green. This scheme depicts the possible joining outcomes. The yellow area represents the gapminder data, the blue the geo data, and the green area the resulting new table.
- With
inner_joinyou join just the rows that match in both tables. So you will not have any new empty values. - The
left_joinandright_joinfunctions are two different perspectives of the same thing, what exactly each does obviously depends on the order in which you feed in the tables. If we always feedgapminderas first (thexargument) andgeosecond (y), left join will keep all rows ingapminderand match them togeo. We know that seven countries will not be matched, which will result in 7 times 12 rows withNAvalues in the new columns containing the abbreviation, main religion in 2008, and the income group. The twelve comes from the number of observations (remember, 1952 to 2007, in five-year intervals). - With
right_join, we will get a monstrous table of allgeo's countries, and the ones that matchgapminder, will be matched twelve times each, with a warning for “many-to-many” relations occurring. The new table is going to have four new columns:year,pop,lifeExp, andgdpPercap. They will be filled withNAexcept in the rows with countries whose names matched across both tables, and the seven countries fromgapminderwill not appear in the new table. - With
full_join, no row will be deleted, but there will beNAin all mismatched rows. - All these tables are going to have the same columns.
gapminder Europe 2007
gapminder_europe <- gapminder %>%
filter(continent == "Europe", year == 2007) %>%
select(!c(continent, year))
glimpse(gapminder_europe)Rows: 30
Columns: 4
$ country <fct> "Albania", "Austria", "Belgium", "Bosnia and Herzegovina", "…
$ lifeExp <dbl> 76.423, 79.829, 79.441, 74.852, 73.005, 75.748, 76.486, 78.3…
$ pop <int> 3600523, 8199783, 10392226, 4552198, 7322858, 4493312, 10228…
$ gdpPercap <dbl> 5937.030, 36126.493, 33692.605, 7446.299, 10680.793, 14619.2…We make a subset of gapminder to keep the data small. Just European countries in 2007 and we drop the columns continent and year because they are all equal.
European subset of geo
Rows: 273
Columns: 5
$ country <chr> "abkh", "abw", "afg", "ago", "aia", "akr_a_dhe", "a…
$ name <chr> "Abkhazia", "Aruba", "Afghanistan", "Angola", "Angu…
$ main_religion_2008 <chr> NA, "christian", "muslim", "christian", "christian"…
$ income_3groups <chr> NA, "high_income", "low_income", "middle_income", N…
$ world_4region <chr> "europe", "americas", "asia", "africa", "americas",…Preferred gapminder, intersection
# A tibble: 29 × 7
country lifeExp pop gdpPercap country.y main_religion_2008 income_3groups
<chr> <dbl> <int> <dbl> <chr> <chr> <chr>
1 Albania 76.4 3.60e6 5937. alb muslim middle_income
2 Austria 79.8 8.20e6 36126. aut christian high_income
3 Belgium 79.4 1.04e7 33693. bel christian high_income
4 Bosnia … 74.9 4.55e6 7446. bih <NA> middle_income
5 Bulgaria 73.0 7.32e6 10681. bgr christian middle_income
6 Croatia 75.7 4.49e6 14619. hrv christian high_income
7 Czech R… 76.5 1.02e7 22833. cze christian high_income
8 Denmark 78.3 5.47e6 35278. dnk christian high_income
9 Finland 79.3 5.24e6 33207. fin christian high_income
10 France 80.7 6.11e7 30470. fra christian high_income
# ℹ 19 more rowsWe would like to add information from geo to gapminder_europe.
The resulting data frame is going to inherit only the rows with matching country names from both.
The _join functions in dplyr always need a first data frame (x), a second data frame (y), and a vector of column names that provide the key pair(s) in the argument called by. The whole thing reads: “Join x with y by this vector of key pairs.” Each pair contains the name of the key column in x and the name of the corresponding column in y, in exactly this order. Relational databases are designed to answer most queries with just one pair, but sometimes you need several to uniquely identify observations in at least one of the data frames.
Mind the quotes in the column names in by. This time they are mandatory.
There were 30 countries in gapminder_europe, but the resulting data frame has only 29 rows. One country was missing in geo_europe, although it is so much longer than gapminder_europe. We will find out later which. You can see an NA in the fourth row, 6th column. This has nothing to do with joining; it was like this in the original geo_europe data frame.
Look closely at the names of the columns. Can you see country and country.y? The former is from gapminder, the latter from geo_europe. Column names must remain unique, therefore dplyr adds suffixes to duplicate names. Typically, the duplicate column name from the x data frame gets the suffix .x and the other one .y. You can replace them with your own suffixes in the suffix argument. In this case, only the one from the y data frame got the suffix. This is because it was used as a by column. By default, the key column from y disappears and only the x key column stays. You can also fiddle with this in a dedicated argument keep. So, in this case, the x country column stayed, the paired name column from y vanished, and then dplyr spotted another country column among the columns inherited from y. The function is just written in such a way that it does only put the suffix on one, when the other one was used as key. Do not ponder on it, just remember.
Keep gapminder intact, no matter what.
One country gets NA in geo_europe columns.
# A tibble: 30 × 7
country lifeExp pop gdpPercap country.y main_religion_2008 income_3groups
<chr> <dbl> <int> <dbl> <chr> <chr> <chr>
1 Albania 76.4 3.60e6 5937. alb muslim middle_income
2 Austria 79.8 8.20e6 36126. aut christian high_income
3 Belgium 79.4 1.04e7 33693. bel christian high_income
4 Bosnia … 74.9 4.55e6 7446. bih <NA> middle_income
5 Bulgaria 73.0 7.32e6 10681. bgr christian middle_income
6 Croatia 75.7 4.49e6 14619. hrv christian high_income
7 Czech R… 76.5 1.02e7 22833. cze christian high_income
8 Denmark 78.3 5.47e6 35278. dnk christian high_income
9 Finland 79.3 5.24e6 33207. fin christian high_income
10 France 80.7 6.11e7 30470. fra christian high_income
# ℹ 20 more rowsWhich is missing?
- country mismatch \(\rightarrow\)
is.na(country.y)
Focus on geo_europe
# A tibble: 73 × 7
country lifeExp pop gdpPercap country.y main_religion_2008 income_3groups
<chr> <dbl> <int> <dbl> <chr> <chr> <chr>
1 Albania 76.4 3.60e6 5937. alb muslim middle_income
2 Austria 79.8 8.20e6 36126. aut christian high_income
3 Belgium 79.4 1.04e7 33693. bel christian high_income
4 Bosnia … 74.9 4.55e6 7446. bih <NA> middle_income
5 Bulgaria 73.0 7.32e6 10681. bgr christian middle_income
6 Croatia 75.7 4.49e6 14619. hrv christian high_income
7 Czech R… 76.5 1.02e7 22833. cze christian high_income
8 Denmark 78.3 5.47e6 35278. dnk christian high_income
9 Finland 79.3 5.24e6 33207. fin christian high_income
10 France 80.7 6.11e7 30470. fra christian high_income
# ℹ 63 more rowsMismatches in geo_europe
dplyr::right_join(x = gapminder_europe, y = geo_europe,
by = c("country" = "name")) %>%
filter(is.na(lifeExp) | is.na(pop) | is.na(gdpPercap))# A tibble: 44 × 7
country lifeExp pop gdpPercap country.y main_religion_2008 income_3groups
<chr> <dbl> <int> <dbl> <chr> <chr> <chr>
1 Abkhazia NA NA NA abkh <NA> <NA>
2 Akrotiri… NA NA NA akr_a_dhe <NA> <NA>
3 Åland NA NA NA ala <NA> <NA>
4 Andorra NA NA NA and christian high_income
5 Armenia NA NA NA arm christian middle_income
6 Antarcti… NA NA NA ata <NA> <NA>
7 Azerbaij… NA NA NA aze muslim middle_income
8 Belarus NA NA NA blr christian middle_income
9 Channel … NA NA NA chanisl christian high_income
10 Czechosl… NA NA NA cheslo <NA> <NA>
# ℹ 34 more rowsWe must look for a column that was not in geo_europe. The country column now is the result of country and name, so it will contain all countries from geo_europe but will have dropped United Kingdom, which was only in gapminder_europe. Any of the numeric columns from gapminder will help us though.
Get what you can
# A tibble: 74 × 7
country lifeExp pop gdpPercap country.y main_religion_2008 income_3groups
<chr> <dbl> <int> <dbl> <chr> <chr> <chr>
1 Albania 76.4 3.60e6 5937. alb muslim middle_income
2 Austria 79.8 8.20e6 36126. aut christian high_income
3 Belgium 79.4 1.04e7 33693. bel christian high_income
4 Bosnia … 74.9 4.55e6 7446. bih <NA> middle_income
5 Bulgaria 73.0 7.32e6 10681. bgr christian middle_income
6 Croatia 75.7 4.49e6 14619. hrv christian high_income
7 Czech R… 76.5 1.02e7 22833. cze christian high_income
8 Denmark 78.3 5.47e6 35278. dnk christian high_income
9 Finland 79.3 5.24e6 33207. fin christian high_income
10 France 80.7 6.11e7 30470. fra christian high_income
# ℹ 64 more rowsThe full join is going to contain all country names from both data frames, with NA where they mismatched.
anti_join detects mismatches instantly
- like
setdiffin vectors
anti_join the other way round
# A tibble: 44 × 4
country name main_religion_2008 income_3groups
<chr> <chr> <chr> <chr>
1 abkh Abkhazia <NA> <NA>
2 akr_a_dhe Akrotiri and Dhekelia <NA> <NA>
3 ala Åland <NA> <NA>
4 and Andorra christian high_income
5 arm Armenia christian middle_income
6 ata Antarctica <NA> <NA>
7 aze Azerbaijan muslim middle_income
8 blr Belarus christian middle_income
9 chanisl Channel Islands christian high_income
10 cheslo Czechoslovakia <NA> <NA>
# ℹ 34 more rowsanti_join: Filter rows of x that are not matched in y.
semi_join detects matches
# A tibble: 29 × 4
country name main_religion_2008 income_3groups
<chr> <chr> <chr> <chr>
1 alb Albania muslim middle_income
2 aut Austria christian high_income
3 bel Belgium christian high_income
4 bgr Bulgaria christian middle_income
5 bih Bosnia and Herzegovina <NA> middle_income
6 che Switzerland christian high_income
7 cze Czech Republic christian high_income
8 deu Germany christian high_income
9 dnk Denmark christian high_income
10 esp Spain christian high_income
# ℹ 19 more rowsFilter rows of x that are matched by y. In this context, the countries are going to be the same, no matter in which order you name the data frames, but you will of course get the rows of the one you fed in as x. It can be helpful to look at both sides, especially when you know that you have a lot of NA across the columns of both. If you have to opt for either left or right join, you can at least choose that with less missing data.
When your observations are not unique
The two tibbles: math
The two tibbles: social sciences
# A tibble: 4 × 3
name birthplace soc_test
<chr> <chr> <dbl>
1 Mary Brown Milan 12
2 John Smith Honolulu 5
3 John Smith Prague 76
4 Helene Field Beijing 49Students’ grades in two courses. You would like to have both exams in the same table. Are the students uniquely identified?
Possible rescue: unique by several columns
No chance to join
If you cannot find anything that makes them unique.
maths2 <- select(maths, -birthplace)
social_sciences2 <- select(social_sciences, !birthplace)
left_join(maths2, social_sciences2, by = "name")Warning in left_join(maths2, social_sciences2, by = "name"): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 1 of `x` matches multiple rows in `y`.
ℹ Row 2 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.# A tibble: 6 × 3
name math_test soc_test
<chr> <dbl> <dbl>
1 John Smith 72 5
2 John Smith 72 76
3 Mary Brown 40 12
4 John Smith 25 5
5 John Smith 25 76
6 Helene Field 91 49Note John Smith occurring four times - all possible combinations get generated.
dplyr::join help
explore the arguments
relationshipmultipleunmatched
Data with typos in the key column(s)
libraries
fuzzyjoinalong withstringdist(used byfuzzyjoin)DataCamp course Intermediate Regular Expressions in R > Similarities Between Strings
You should only know that there are ways to cope with strings in columns that do not match completely. Only read further if you are particularly interested.
JRC Names
Steinberger Ralf, Bruno Pouliquen, Mijail Kabadjov, Jenya Belyaeva & Erik van der Goot (2011).JRC-Names: A freely available, highly multilingual named entity resource. Proceedings of the 8th International Conference Recent Advances in Natural Language Processing (RANLP). Hissar, Bulgaria, 12-14 September 2011.
JRC Person Names
- persons with 4 spellings of their names
- 2 tables, 20 rows,
- equal pers IDs, different spelling
- test different fuzzy join metrics
Watch the number of rows in inner join. Sensitivity (recall): how many it catches of those to catch: the longer table the better Specificity (precision): how many of those caught were correct (how much noise): check that their ids match.
JRC each table
# A tibble: 5 × 2
id name
<dbl> <chr>
1 41 John+Ashcroft
2 46 Richard+Boucher
3 56 Adam+Ereli
4 92 Chris+Patten
5 123 Dan+Senor Matching on Levenschtein Distance
joinJRC_lv <- fuzzyjoin::stringdist_inner_join(x = jrc_1, y = jrc_2,
distance_col = "distance", by = "name", ignore_case = TRUE,
method = "lv", max_dist = 2) %>%
relocate(name.x, name.y, distance) %>% select(!c(n.x, n.y, index_id.x, starts_with("PersOrg")))
joinJRC_lv# A tibble: 17 × 6
name.x name.y distance id.x id.y index_id.y
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 John+Ashcroft John+Ascroft 1 41 41 2
2 Adam+Ereli Adam+J+Ereli 2 56 56 2
3 Chris+Patten CHRIS+PATTEN 0 92 92 2
4 Peter+Hain PETER+HAIN 0 159 159 2
5 Roberto+Castelli Robero+Castelli 1 173 173 2
6 Shaukat+Sultan Shaukat+Sultán 1 174 174 2
7 Gerhard+Mayer+Vorfelder Gerhard+Mayer-Vorfel… 1 196 196 2
8 Johannes+Rau JOHANNES+RAU 0 202 202 2
9 Roland+Koch ROLAND+KOCH 0 215 215 2
10 Jesus+Caldera Jesús+Caldera 1 231 231 2
11 Martin+Bartenstein Martin+Barteinstein 1 241 241 2
12 Klaus+Zumwinkel Klaus+Zumwinckel 1 252 252 2
13 Philip+Green Phillip+Green 1 253 253 2
14 Umberto+Agnelli UMBERTO+AGNELLI 0 259 259 2
15 Marco+Follini MARCO+FOLLINI 0 284 284 2
16 Bernard+Thibault BERNARD+THIBAULT 0 292 292 2
17 Seamus+Brennan Séamus+Brennan 1 339 339 2Matching on cosine distance between qgrams
joinJRC12_cosine <- fuzzyjoin::stringdist_inner_join(x = jrc_1, y = jrc_2, distance_col = "distance",
by = "name", ignore_case = TRUE,
method = "cosine",
q = 1,
max_dist = 0.15,
) %>% relocate(name.x, name.y, distance) %>% select(!c(n.x, n.y, index_id.x, starts_with("PersOrg")))
joinJRC12_cosine# A tibble: 20 × 6
name.x name.y distance id.x id.y index_id.y
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 John+Ashcroft John+Ascroft 0.0277 41 41 2
2 Richard+Boucher Rick+Boucher 0.1 46 46 2
3 Adam+Ereli Adam+J+Ereli 0.0551 56 56 2
4 Chris+Patten CHRIS+PATTEN 0 92 92 2
5 Dan+Senor Daniel+Senor 0.0955 123 123 2
6 Peter+Hain PETER+HAIN 0 159 159 2
7 Roberto+Castelli Robero+Castelli 0.0186 173 173 2
8 Shaukat+Sultan Shaukat+Sultán 0.0383 174 174 2
9 Gerhard+Mayer+Vorfelder Gerhard+Mayer-Vorfel… 0.0165 196 196 2
10 Johannes+Rau JOHANNES+RAU 0 202 202 2
11 Roland+Koch ROLAND+KOCH 0 215 215 2
12 Jesus+Caldera Jesús+Caldera 0.0526 231 231 2
13 Martin+Bartenstein Martin+Barteinstein 0.0105 241 241 2
14 Klaus+Zumwinkel Klaus+Zumwinckel 0.0230 252 252 2
15 Philip+Green Phillip+Green 0.0227 253 253 2
16 Umberto+Agnelli UMBERTO+AGNELLI 0 259 259 2
17 Thomas+Kean Tom+Kean 0.117 274 274 2
18 Marco+Follini MARCO+FOLLINI 0 284 284 2
19 Bernard+Thibault BERNARD+THIBAULT 0 292 292 2
20 Seamus+Brennan Séamus+Brennan 0.0392 339 339 2Matching on Jaccard distance
joinJRC12_jaccard <- fuzzyjoin::stringdist_inner_join(x = jrc_1, y = jrc_2,
distance_col = "distance", by = "name", ignore_case = TRUE, method = "jaccard",
q = 1, max_dist = 0.18) %>%
relocate(name.x, name.y, distance) %>% select(!c(n.x, n.y, index_id.x, starts_with("PersOrg")))
joinJRC12_jaccard# A tibble: 16 × 6
name.x name.y distance id.x id.y index_id.y
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 John+Ashcroft John+Ascroft 0 41 41 2
2 Adam+Ereli Adam+J+Ereli 0.111 56 56 2
3 Chris+Patten CHRIS+PATTEN 0 92 92 2
4 Peter+Hain PETER+HAIN 0 159 159 2
5 Roberto+Castelli Robero+Castelli 0 173 173 2
6 Shaukat+Sultan Shaukat+Sultán 0.1 174 174 2
7 Gerhard+Mayer+Vorfelder Gerhard+Mayer-Vorfel… 0.0714 196 196 2
8 Johannes+Rau JOHANNES+RAU 0 202 202 2
9 Roland+Koch ROLAND+KOCH 0 215 215 2
10 Martin+Bartenstein Martin+Barteinstein 0 241 241 2
11 Klaus+Zumwinkel Klaus+Zumwinckel 0.0769 252 252 2
12 Philip+Green Phillip+Green 0 253 253 2
13 Umberto+Agnelli UMBERTO+AGNELLI 0 259 259 2
14 Marco+Follini MARCO+FOLLINI 0 284 284 2
15 Bernard+Thibault BERNARD+THIBAULT 0 292 292 2
16 Seamus+Brennan Séamus+Brennan 0.1 339 339 2